Tomography subpackage (CHAP.tomo)
The tomography subpackage contains the modules that are unique to tomography data processing workflows. This document describes how to run a tomography reconstruction workflow in a Linux terminal.
Tomographic reconstruction
Tomography is a type of 3D imaging that uses some type of penetrative wave (an X-ray beam at CHESS). Tomographic reconstruction refers to the process of recovering 3D spatial information on an object from a set of projected images acquired under different angles after transmission of the beam through the sample.
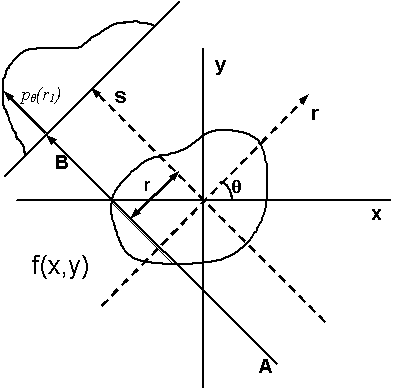
Illustration of tomographic reconstruction (https://en.wikipedia.org/wiki/Tomographic_reconstruction). The projected image on the detector results from transmission of the beam through the sample at a given angle at varying locations. The intensity at the detector depends on the amount of scattering and absorption in the sample or on, what is often called, its linear attenuation coefficient. Spatial variations in for example density can lead to spatial variations of the local attenuation coefficient. The intensity on the detector is basically a measure of its line integral along a path \(AB\) at a projected sample position \(r\). We can then obtain full 3D reconstruction of the spatial variation from a set of 2D images at varying angles \(\theta\) by a mathematical inversion technique called the inverse Radon transform (https://en.wikipedia.org/wiki/Radon_transform).
The input data
A standard tomographic experiment at CHESS (and other similar institutions) consist of a set of X-ray transmission measurements captured on a 2D area detector. It typically involves two (sets of) measurements without a sample in the beam, one with the beam stop in place (the dark image), and one with the beam stop open (the bright image). Followed by a measurement of a stack of images (a tomographic image series) at varying rotation angle (over 180 or a full 360 degrees of rotation) with the sample in the beam secured to a rotating sample holder. Note that if the sample is larger than the beam cross section, this may require multiple image stacks at various sample positions in the plane perpendicular to the X-ray beam.
Processing the data
A standard tomographic reconstruction in CHAP consists of three steps:
Reducing the data, i.e., correcting the raw detector images for background and non-uniformities in the beam intensity profile using dark and bright fields collected separately from the tomography image series.
Finding the calibrated rotation axis. Accurate reconstruction relies on accurately knowing the center of rotation at each data plane perpendicular to the rotation axis (the sinogram). This rotation axis is calibrated by selecting two data planes, one near the top and one near the bottom of the sample or beam, and visually or automatically picking the optimal center location.
Reconstructing the reduced data for the calibrated rotation axis. For samples taller than the height of the beam, this last step can consist of two parts:
reconstruction of each individual stack of images, and
combining the individual stacks into one 3D reconstructed data set.
Note that combining stacks with a horizontal displacement for samples wider than the width of the beam is not yet implemented.
Activating the tomography conda environment
From the CHESS Compute Farm
Log in to the CHESS Compute Farm and activate the CHAP_tomo environment:
source /nfs/chess/sw/miniforge3_chap/bin/activate
conda activate CHAP_tomo
From a local CHAP clone
Create and activate a base conda environent, e.g. with Miniforge.
Install a local version of the CHAP package according to the installation instructions.
Create the tomography conda environment:
mamba env create -f <path_to_CHAP_clone_dir>/CHAP/tomo/environment.yml
Activate the
CHAP_tomoenvironment:conda activate CHAP_tomo
Running a tomography reconstruction
Navigate to your work directory.
Create the required CHAP pipeline file for the workflow (see below) and any additional workflow specific input files. This typically includes at a minimum the detector configuration
.yamlfile with the detector image size information. For example, for the andor2 detector (andor2.yaml):prefix: andor2 rows: 2160 columns: 2560 pixel_size: - 0.0065 - 0.0065 lens_magnification: 5.0
Here, the prefix field must equal a detector ID field in the
detectorslist in the pipeline input file.Run the reconstruction:
CHAP <pipelinefilename>Respond to any prompts that pop up if running interactively.
Inspecting output
The output consists of a single NeXus (.nxs) file containing the reconstructed data set and a metadata record that can be processed and saved or uploaded to an external metadata service downstream in the pipeline file. Additionally, optional output figures (.png) may be saved to an output directory specified in the pipeline file.
The optional output figures can be viewed directly by any PNG image viewer. The data in the NeXus output file can be viewed in NeXpy, a high-level python interface to HDF5 files, particularly those stored as NeXus data:
Open the NeXpy GUI by entering in your terminal:
nexpy &
After the GUI pops up, click File -> Open to navigate to the folder where your output
.nxsfile was saved, and select it.Double click on the base level
NXrootfield in the leftmost “NeXus Data” panel to view the reconstruction. Note that theNXrootname is always the basename of the output file.Or navigate the filetree in the “NeXus Data” panel to inspect any other output or metadata field. Note that the latest data set in any tomography reconstruction workflow is always available under the “data”
NXdatafield among the defaultNXentry’s fields (it is this data set that is opened in the viewer panel when double clicking theNXrootfield). The defaultNXentryname is always the “title” field in the workflow’s map configuration.
Creating the pipeline file
Create a workflow pipeline.yaml file according to the CHAP pipeline instructions. A generic pipeline input file for a full tomography reconstruction workflow is as follows (note that spaces and indentation are important in .yaml files):
config:
root: . # Change as desired
inputdir: . # Change as desired
# Path can be relative to root (line 2) or absolute
outputdir: output # Change as desired
# Path can be relative to root (line 2) or absolute
interactive: true # Change as desired
log_level: info # Set to debug, info, warning, or error
pipeline:
# Create the map
- common.MapProcessor:
config:
title: <your_BTR> # Change as desired, typically BTR
station: id3a # Change as needed
experiment_type: TOMO
sample:
name: <your_sample_name> # Change as desired
# typically the sample name
spec_scans: # Edit both SPEC log file path and tomography scan numbers
# Path can be relative to inputdir (line 3) or absolute
- spec_file: <your_raw_data_directory>/spec.log
scan_numbers: [3, 4, 5]
independent_dimensions:
- label: rotation_angles
units: degrees
data_type: scan_column
name: theta
- label: x_translation
units: mm
data_type: smb_par
name: ramsx # Change as needed
- label: z_translation
units: mm
data_type: smb_par
name: ramsz # Change as needed
detector_config:
detectors:
- id: andor2 # Change as needed
schema: tomofields
- common.SpecReader:
config:
station: id3a # Change as needed
experiment_type: TOMO
spec_scans: # Edit both SPEC log file path and tomography scan numbers
# Path can be relative to inputdir (line 3) or absolute
- spec_file: <your_raw_data_directory>/spec.log
scan_numbers: 1
detector_config:
detectors:
- id: andor2 # Change as needed
schema: darkfield
- common.SpecReader:
config:
station: id3a # Change as needed
experiment_type: TOMO
spec_scans: # Edit both SPEC log file path and tomography scan numbers
# Path can be relative to inputdir (line 3) or absolute
- spec_file: <your_raw_data_directory>/spec.log
scan_numbers: 2
detector_config:
detectors:
- id: andor2 # Change as needed
schema: brightfield
- common.YAMLReader:
filename: andor2.yaml # Detector config file
# Path can be relative to inputdir (line 3) or absolute
schema: tomo.models.Detector
- tomo.TomoCHESSMapConverter
# Full tomography reconstruction
- tomo.TomoDataProcessor:
reduce_data: true
find_center: true
reconstruct_data: true
combine_data: true # Only needed for a stack of tomography image sets
save_figures: true
- common.NexusWriter:
filename: reconstructed_sample.nxs # Change as desired
# will be placed in 'outdutdir' (line 5)
force_overwrite: true # Do not set to false!
# Rename an existing file if you want to prevent
# it from being overwritten
- common.ImageWriter:
outputdir: figures # Change as desired, unless an absolute path
# this will appear under 'outdutdir' (line 5)
force_overwrite: true # Do not set to false!
# Rename an existing file if you want to prevent
# it from being overwritten
Example
The CHAP tomography subpackage comes with several workflow examples, one of them for an CHESS ID3A beamline style experiment of a truncated hollow four sided pyramid made from a single homogeneous material.
This example uses simulated raw imaging data that needs to be available in a specific location ahead of the reconstruction. If you are logged in on the CHESS Compute Farm, replace <path_to_CHAP_clone_dir> below with /nfs/chess/sw/ChessAnalysisPipeline, the path to the CHAP repository administrated by the CHAP developers. If not, replace it with the path to your local CHAP repository. In the later case, you will also need to create the raw data once, since it is not part of the cloned repository. To do so, create and activate your local CHAP_tomo conda environment as instructed above, navigate to your local CHAP repository, and execute:
CHAP examples/tomo/pipeline_id3a_pyramid_sim.yaml
To perform the reconstruction:
Create a work directory in your own user space.
Within the work directory, create a plain text file, named
pipeline_id3a_pyramid.yaml, with the following content (note that spaces and indentation are important in.yamlfiles):config: root: . inputdir: <path_to_CHAP_clone_dir>/examples/tomo/config outputdir: hollow_pyramid interactive: true log_level: info pipeline: # Create the map - common.MapProcessor: config: title: hollow_pyramid station: id3a experiment_type: TOMO sample: name: hollow_pyramid spec_scans: - spec_file: ../data/hollow_pyramid/spec.log scan_numbers: [3, 4, 5] independent_dimensions: - label: rotation_angles units: degrees data_type: scan_column name: theta - label: x_translation units: mm data_type: smb_par name: ramsx - label: z_translation units: mm data_type: smb_par name: ramsz detector_config: detectors: - id: sim schema: tomofields - common.SpecReader: config: station: id3a experiment_type: TOMO spec_scans: - spec_file: ../data/hollow_pyramid/spec.log scan_numbers: 1 detector_config: detectors: - id: sim schema: darkfield - common.SpecReader: config: station: id3a experiment_type: TOMO spec_scans: - spec_file: spec.log scan_numbers: 2 detector_config: detectors: - id: sim schema: brightfield - common.YAMLReader: filename: detector_pyramid.yaml schema: tomo.models.Detector - tomo.TomoCHESSMapConverter # Full tomography reconstruction - tomo.TomoDataProcessor: reduce_data: true find_center: true reconstruct_data: true combine_data: true outputdir: saved_figs save_figs: 'only' - common.NexusWriter: filename: combined_hollow_pyramid.nxs force_overwrite: true - common.ImageWriter: outputdir: figures force_overwrite: true
Execute
CHAP pipeline_id3a_pyramid.yamlFollow the interactive prompts or replace
truewithfalseon line 5 inpipeline_id3a_pyramid.yaml(interactive: false) and run the workflow non-interactively.Inspect the results:
In NeXpy, as instructed above, navigate to
<your_work_directory>/hollow_pyramidand opencombined_hollow_pyramid.nxsBy displaying the output figures in
<your_work_directory>/hollow_pyramid/figures
The “config” block defines the CHAP generic configuration parameters:
root: The work directory, defaults to the current directory (whereCHAP <pipelinefilename>is executed). Must be an absolute path or relative to the current directory.inputdir: The default directory for files read by any CHAP reader (must have read access), defaults toroot. Must be an absolute path or relative toroot.outputdir: The default directory for files written by any CHAP writter (must have write access, will be created if not existing), defaults toroot. Must be an absolute path or relative toroot.interactive: Allows for user interactions, defaults tofalse.log_level: The Python logging level.
The “pipeline” block creates the actual workflow pipeline, it this example it consists of six toplevel processes that get executed successively:
common.MapProcessor: A processor that creates a CHESS style map.pipeline.MultiplePipelineItem: A processor that executes (in this case reads) three items and passes the inputs on to the next item in the pipeline.tomo.TomoCHESSMapConverter: A processor that converts the inputs to a CHESS style map.tomo.TomoDataProcessor: The actual tomographic reconstruction processor that creates a single NeXus object with the reconstructed data as well as all metadata pertaining to the reconstruction and passes it on to the next item in the pipeline.common.NexusWriter: A processor that writes the reconstructed data to a NeXus file.common.ImageWriter: A pocessor that writes any output figures created bytomo.TomoDataProcessorto a directoryfiguresunderneath the workflow output directory.